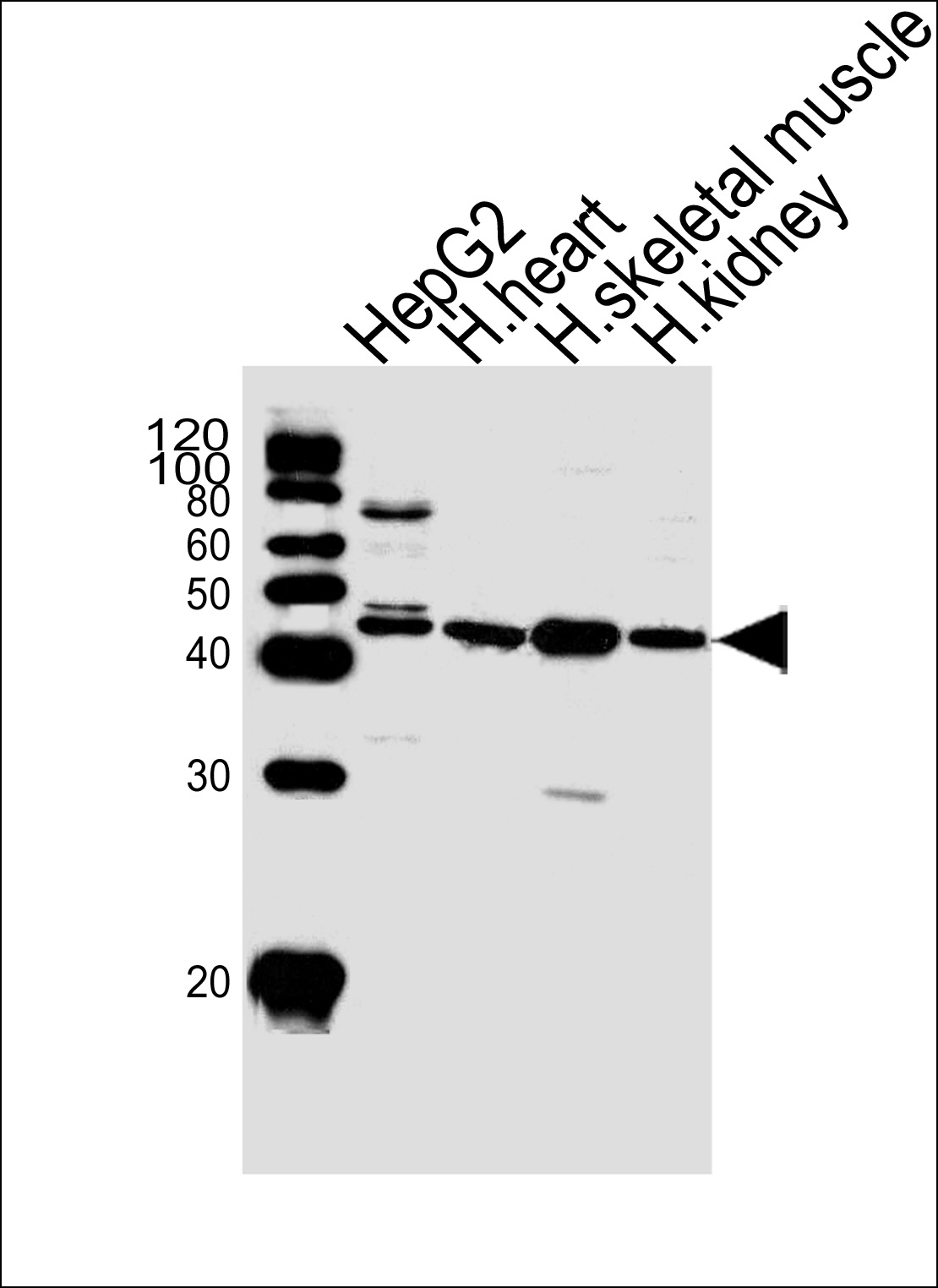
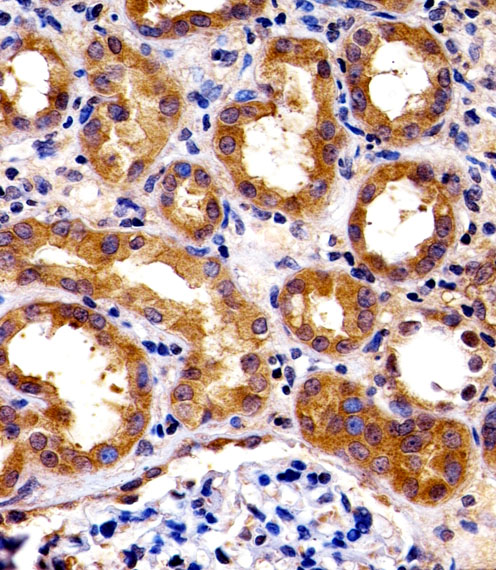
SPHK1 Antibody (N-term P74)
Purified Rabbit Polyclonal Antibody (Pab)
- SPECIFICATION
- CITATIONS: 2
- PROTOCOLS
- BACKGROUND

Application
| IHC-P, WB, E |
|---|---|
| Primary Accession | Q9NYA1 |
| Reactivity | Human |
| Host | Rabbit |
| Clonality | Polyclonal |
| Isotype | Rabbit IgG |
| Calculated MW | 42518 Da |
| Antigen Region | 59-89 aa |
| Gene ID | 8877 |
|---|---|
| Other Names | Sphingosine kinase 1, SK 1, SPK 1, SPHK1, SPHK, SPK |
| Target/Specificity | This SPHK1 antibody is generated from rabbits immunized with a KLH conjugated synthetic peptide between 59-89 amino acids from the N-terminal region of human SPHK1. |
| Dilution | WB~~1:1000 IHC-P~~1:100 |
| Format | Purified polyclonal antibody supplied in PBS with 0.09% (W/V) sodium azide. This antibody is prepared by Saturated Ammonium Sulfate (SAS) precipitation followed by dialysis against PBS. |
| Storage | Maintain refrigerated at 2-8°C for up to 2 weeks. For long term storage store at -20°C in small aliquots to prevent freeze-thaw cycles. |
| Precautions | SPHK1 Antibody (N-term P74) is for research use only and not for use in diagnostic or therapeutic procedures. |
| Name | SPHK1 (HGNC:11240) |
|---|---|
| Function | Catalyzes the phosphorylation of sphingosine to form sphingosine 1-phosphate (SPP), a lipid mediator with both intra- and extracellular functions. Also acts on D-erythro-sphingosine and to a lesser extent sphinganine, but not other lipids, such as D,L-threo- dihydrosphingosine, N,N-dimethylsphingosine, diacylglycerol, ceramide, or phosphatidylinositol (PubMed:20577214, PubMed:23602659, PubMed:29662056, PubMed:24929359, PubMed:11923095). In contrast to proapoptotic SPHK2, has a negative effect on intracellular ceramide levels, enhances cell growth and inhibits apoptosis (PubMed:16118219). Involved in the regulation of inflammatory response and neuroinflammation. Via the product sphingosine 1-phosphate, stimulates TRAF2 E3 ubiquitin ligase activity, and promotes activation of NF- kappa-B in response to TNF signaling leading to IL17 secretion (PubMed:20577214). In response to TNF and in parallel to NF-kappa-B activation, negatively regulates RANTES induction through p38 MAPK signaling pathway (PubMed:23935096). Involved in endocytic membrane trafficking induced by sphingosine, recruited to dilate endosomes, also plays a role on later stages of endosomal maturation and membrane fusion independently of its kinase activity (PubMed:28049734, PubMed:24929359). In Purkinje cells, seems to be also involved in the regulation of autophagosome-lysosome fusion upon VEGFA (PubMed:25417698). |
| Cellular Location | Cytoplasm. Nucleus. Cell membrane. Endosome membrane; Peripheral membrane protein. Membrane, clathrin-coated pit. Synapse {ECO:0000250|UniProtKB:Q8CI15} Note=Translocated from the cytoplasm to the plasma membrane in a CIB1- dependent manner (PubMed:19854831). Binds to membranes containing negatively charged lipids but not neutral lipids (PubMed:24929359) Recruited to endocytic membranes by sphingosine where promotes membrane fusion (By similarity). {ECO:0000250|UniProtKB:Q8CI15, ECO:0000269|PubMed:19854831, ECO:0000269|PubMed:24929359} |
| Tissue Location | Widely expressed with highest levels in adult liver, kidney, heart and skeletal muscle. Expressed in brain cortex (at protein level) (PubMed:29662056). |
Provided below are standard protocols that you may find useful for product applications.
Background
Sphingosine Kinase (SphK) catalyzes the phosphorylation of the lipid sphingosine, creating the bioactive lipid sphingosine-1-phosphate (S1P). S1P subsequently signals through cell surface G protein-coupled receptors, as well as intracellularly, to modulate cell proliferation, survival, motility and differentiation. SphK is an important signaling enzyme which is activated by diverse agents, including growth factors that signal through receptor tyrosine kinases, agents activating G protein-coupled receptors, and immunoglobulin receptors. Two SphK isotypes, SphK-1 and SphK-2, have been cloned, and both isotypes are ubiquitously expressed. SphK-1 has been shown to mediate cell growth, prevention of apoptosis, and cellular transformation, and is upregulated in a variety of human tumors. In contrast, SphK-2 increases apoptosis, and may be responsible for phosphorylating and activating the immunosuppressive drug FTY720.
References
Ota, T., et al., Nat. Genet. 36(1):40-45 (2004).
Nava, V.E., et al., FEBS Lett. 473(1):81-84 (2000).
Melendez, A.J., et al., Gene 251(1):19-26 (2000).
Pitson, S.M., et al., Biochem. J. 350 Pt 2, 429-441 (2000).
If you have used an Abcepta product and would like to share how it has performed, please click on the "Submit Review" button and provide the requested information. Our staff will examine and post your review and contact you if needed.
If you have any additional inquiries please email technical services at tech@abcepta.com.





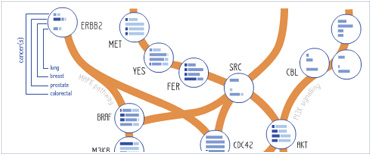







 Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them.
Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them. The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle.
The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle. The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.
The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.